Smile
Smile (Statistical Machine Intelligence and Learning Engine) is a fast and comprehensive machine learning, NLP, linear algebra, graph, interpolation, and visualization system in Java and Scala. With advanced data structures and algorithms, Smile delivers state-of-art performance. Smile is well documented and please check out the project website for programming guides and more information.
Smile covers every aspect of machine learning, including classification, regression, clustering, association rule mining, feature selection, manifold learning, multidimensional scaling, genetic algorithms, missing value imputation, efficient nearest neighbor search, etc.
Smile implements the following major machine learning algorithms:
-
Classification: Support Vector Machines, Decision Trees, AdaBoost, Gradient Boosting, Random Forest, Logistic Regression, Neural Networks, RBF Networks, Maximum Entropy Classifier, KNN, Naïve Bayesian, Fisher/Linear/Quadratic/Regularized Discriminant Analysis.
-
Regression: Support Vector Regression, Gaussian Process, Regression Trees, Gradient Boosting, Random Forest, RBF Networks, OLS, LASSO, ElasticNet, Ridge Regression.
-
Feature Selection: Genetic Algorithm based Feature Selection, Ensemble Learning based Feature Selection, TreeSHAP, Signal Noise ratio, Sum Squares ratio.
-
Clustering: BIRCH, CLARANS, DBSCAN, DENCLUE, Deterministic Annealing, K-Means, X-Means, G-Means, Neural Gas, Growing Neural Gas, Hierarchical Clustering, Sequential Information Bottleneck, Self-Organizing Maps, Spectral Clustering, Minimum Entropy Clustering.
-
Association Rule & Frequent Itemset Mining: FP-growth mining algorithm.
-
Manifold Learning: IsoMap, LLE, Laplacian Eigenmap, t-SNE, UMAP, PCA, Kernel PCA, Probabilistic PCA, GHA, Random Projection, ICA.
-
Multi-Dimensional Scaling: Classical MDS, Isotonic MDS, Sammon Mapping.
-
Nearest Neighbor Search: BK-Tree, Cover Tree, KD-Tree, SimHash, LSH.
-
Sequence Learning: Hidden Markov Model, Conditional Random Field.
-
Natural Language Processing: Sentence Splitter and Tokenizer, Bigram Statistical Test, Phrase Extractor, Keyword Extractor, Stemmer, POS Tagging, Relevance Ranking
You can use the libraries through Maven central repository by adding the following to your project pom.xml file.
<dependency>
<groupId>com.github.haifengl</groupId>
<artifactId>smile-core</artifactId>
<version>2.6.0</version>
</dependency>
For NLP, use the artifactId smile-nlp.
For Scala API, please use
libraryDependencies += "com.github.haifengl" %% "smile-scala" % "2.6.0"
For Kotlin API, add the below into the dependencies section of Gradle build script.
implementation("com.github.haifengl:smile-kotlin:2.6.0")
For Clojure API, add the following dependency to your project or build file:
[org.clojars.haifengl/smile "2.6.0"]
Some algorithms rely on BLAS and LAPACK (e.g. manifold learning, some clustering algorithms, Gaussian Process regression, MLP, etc). To use these algorithms, you should include OpenBLAS for optimized matrix computation:
libraryDependencies ++= Seq(
"org.bytedeco" % "javacpp" % "1.5.4" classifier "macosx-x86_64" classifier "windows-x86_64" classifier "linux-x86_64" classifier "linux-arm64" classifier "linux-ppc64le" classifier "android-arm64" classifier "ios-arm64",
"org.bytedeco" % "openblas" % "0.3.10-1.5.4" classifier "macosx-x86_64" classifier "windows-x86_64" classifier "linux-x86_64" classifier "linux-arm64" classifier "linux-ppc64le" classifier "android-arm64" classifier "ios-arm64",
"org.bytedeco" % "arpack-ng" % "3.7.0-1.5.4" classifier "macosx-x86_64" classifier "windows-x86_64" classifier "linux-x86_64" classifier "linux-arm64" classifier "linux-ppc64le"
)
In this example, we include all supported 64-bit platforms and filter out 32-bit platforms. The user should include only the needed platforms to save spaces.
If you prefer other BLAS implementations, you can use any library found on the "java.library.path" or on the class path, by specifying it with the "org.bytedeco.openblas.load" system property. For example, to use the BLAS library from the Accelerate framework on Mac OS X, we can pass options such as -Djava.library.path=/usr/lib/ -Dorg.bytedeco.openblas.load=blas.
For a default installation of MKL that would be -Dorg.bytedeco.openblas.load=mkl_rt. Or you may simply include smile-mkl module in your project, which includes MKL binaries. With smile-mkl module in the class path, Smile will automatically switch to MKL.
libraryDependencies += "com.github.haifengl" %% "smile-mkl" % "2.6.0"
Shell
Smile comes with interactive shells for Java, Scala and Kotlin. Download pre-packaged Smile from the releases page. In the home directory of Smile, type
./bin/smile
to enter the Scala shell. You can run any valid Scala expressions in the shell. In the simplest case, you can use it as a calculator. Besides, all high-level Smile operators are predefined in the shell. By default, the shell uses up to 75% memory. If you need more memory to handle large data, use the option -J-Xmx or -XX:MaxRAMPercentage. For example,
./bin/smile -J-Xmx30G
You can also modify the configuration file ./conf/smile.ini for the memory and other JVM settings.
To use Java's JShell, type
./bin/jshell.sh
which has Smile's jars in the classpath. Similarly, run
./bin/kotlin.sh
to enter Kotlin REPL.
Model Serialization
Most models support the Java Serializable interface (all classifiers do support Serializable interface) so that you can use them in Spark. For reading/writing the models in non-Java code, we suggest [XStream] (https://github.com/x-stream/xstream) to serialize the trained models. XStream is a simple library to serialize objects to XML and back again. XStream is easy to use and doesn't require mappings (actually requires no modifications to objects). Protostuff is a nice alternative that supports forward-backward compatibility (schema evolution) and validation. Beyond XML, Protostuff supports many other formats such as JSON, YAML, protobuf, etc.
Visualization
Smile provides a Swing-based data visualization library SmilePlot, which provides scatter plot, line plot, staircase plot, bar plot, box plot, histogram, 3D histogram, dendrogram, heatmap, hexmap, QQ plot, contour plot, surface, and wireframe.
To use SmilePlot, add the following to dependencies
<dependency>
<groupId>com.github.haifengl</groupId>
<artifactId>smile-plot</artifactId>
<version>2.6.0</version>
</dependency>
Smile also support data visualization in declarative approach. With smile.plot.vega package, we can create a specification that describes visualizations as mappings from data to properties of graphical marks (e.g., points or bars). The specification is based on Vega-Lite. The Vega-Lite compiler automatically produces visualization components including axes, legends, and scales. It then determines properties of these components based on a set of carefully designed rules.
Gallery
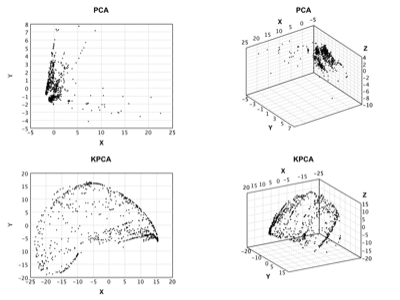 Kernel PCA |
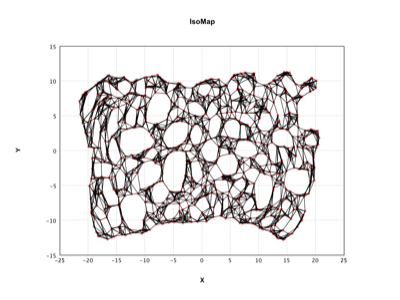 IsoMap |
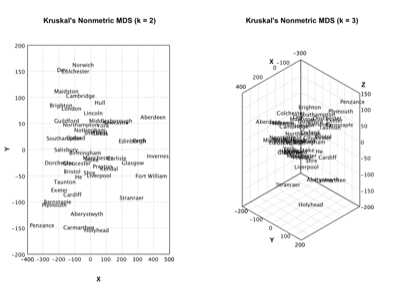 Multi-Dimensional Scaling |
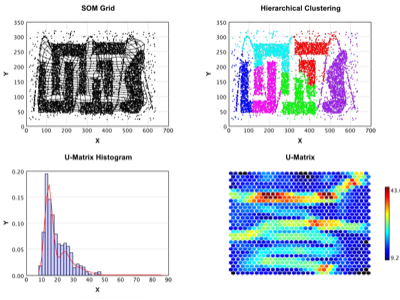 SOM |
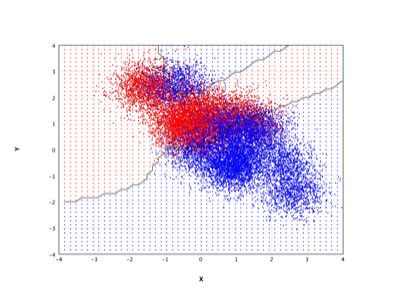 Neural Network |
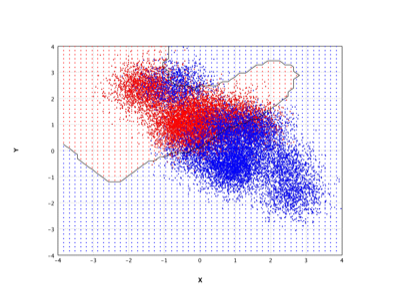 SVM |
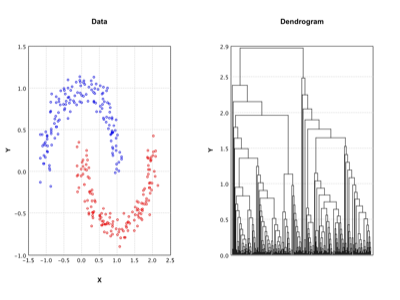 Agglomerative Clustering |
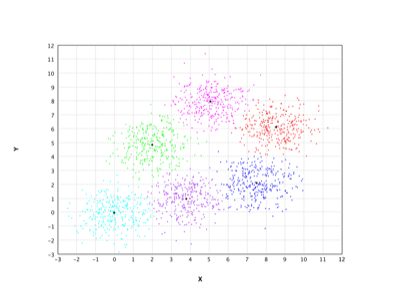 X-Means |
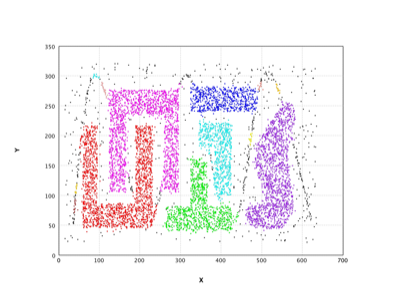 DBSCAN |
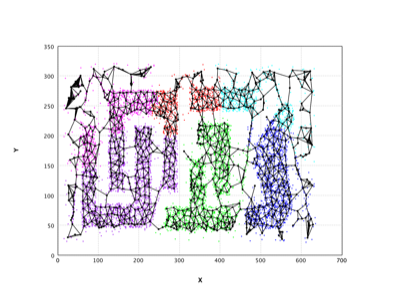 Neural Gas |
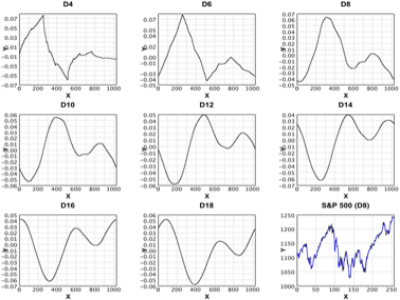 Wavelet |
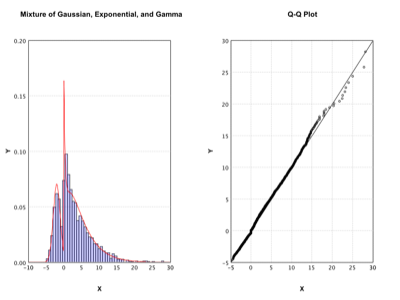 Exponential Family Mixture |
